3.2.3 Paper histology subsets
3.2.3.1 STLMS
There are 36886 genes and 53 samples in the dataset.
At least some samples or genes look not suitable for the co-expression analysis. After removing them, there remain 36778 genes and 53 samples.
## Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
## 1 1 0.330 -3.94 0.960 5160.000 5.06e+03 8280.0
## 2 3 0.730 -3.00 0.986 328.000 2.86e+02 1150.0
## 3 5 0.917 -2.50 0.993 44.900 3.07e+01 354.0
## 4 7 0.971 -2.13 0.984 10.300 4.79e+00 206.0
## 5 9 0.966 -1.80 0.971 3.480 9.75e-01 140.0
## 6 11 0.960 -1.58 0.966 1.590 2.40e-01 103.0
## 7 13 0.942 -1.45 0.947 0.897 6.84e-02 78.8
## 8 15 0.984 -1.32 0.984 0.577 2.16e-02 62.2
## 9 17 0.981 -1.28 0.980 0.406 7.28e-03 52.8
## 10 19 0.935 -1.30 0.931 0.303 2.62e-03 48.2
## 11 21 0.901 -1.33 0.896 0.237 9.84e-04 44.6
## 12 23 0.894 -1.33 0.884 0.192 3.83e-04 41.8
## 13 25 0.881 -1.33 0.863 0.160 1.53e-04 39.5
## 14 27 0.861 -1.36 0.844 0.136 6.30e-05 37.5
## 15 29 0.859 -1.35 0.835 0.118 2.60e-05 35.9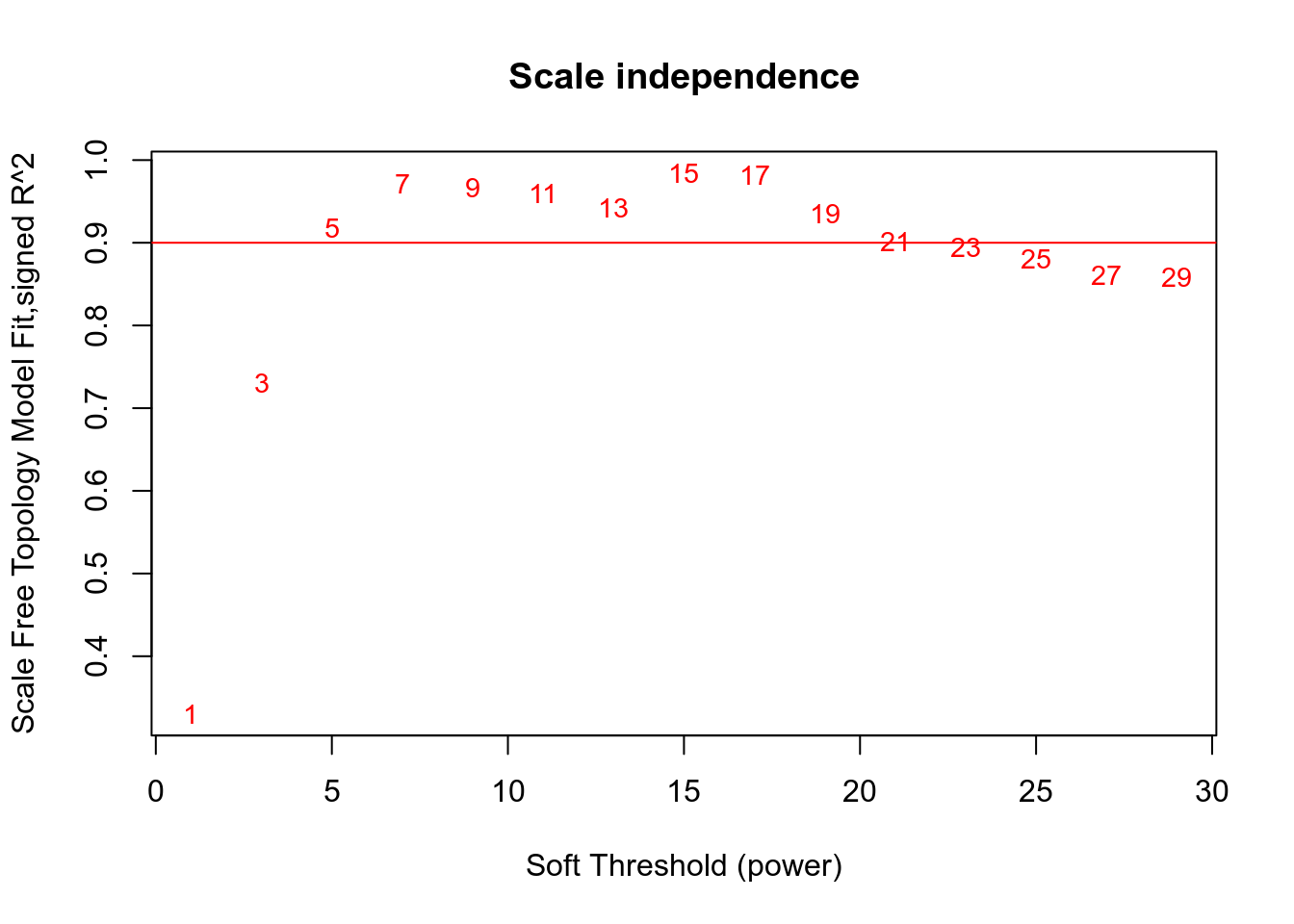
Network is computed using power=5.
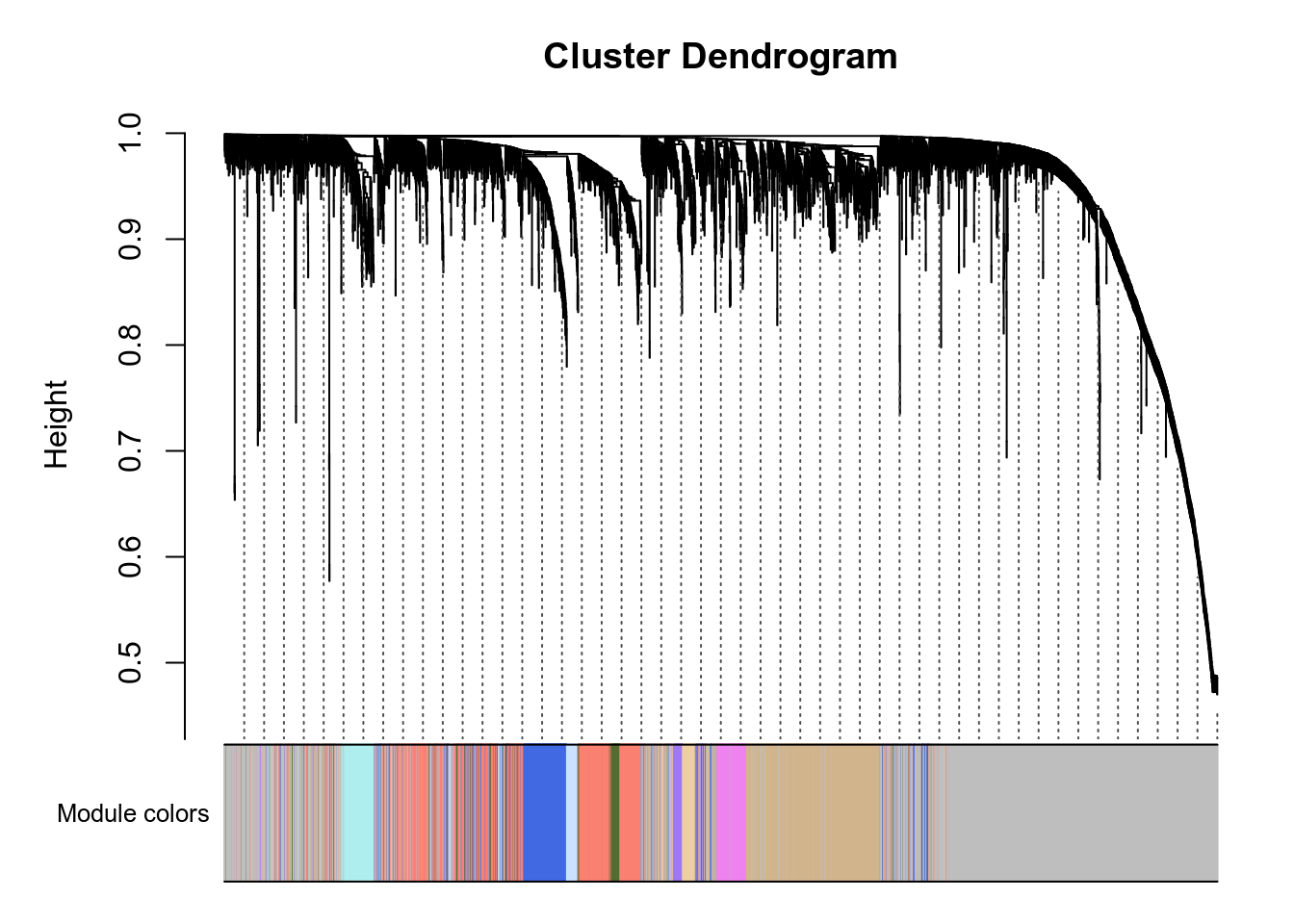
Here are the modules for a few genes of interest:
- VNN1 is in the blue module.
- PPARA is in the tan module.
- PPARD is in the paleturquoise module.
- PPARG is in the blue module.
- PPARGC1A is in the green module.
- PPARGC1B is in the violet module.
g:Profiler results for all modules are available here. Since there are many modules, you should expect at least 1 minute computation on the g:Profiler website for this query. Check below for faster queries focused on VNN1 module.

Figure 3.117: Pathway
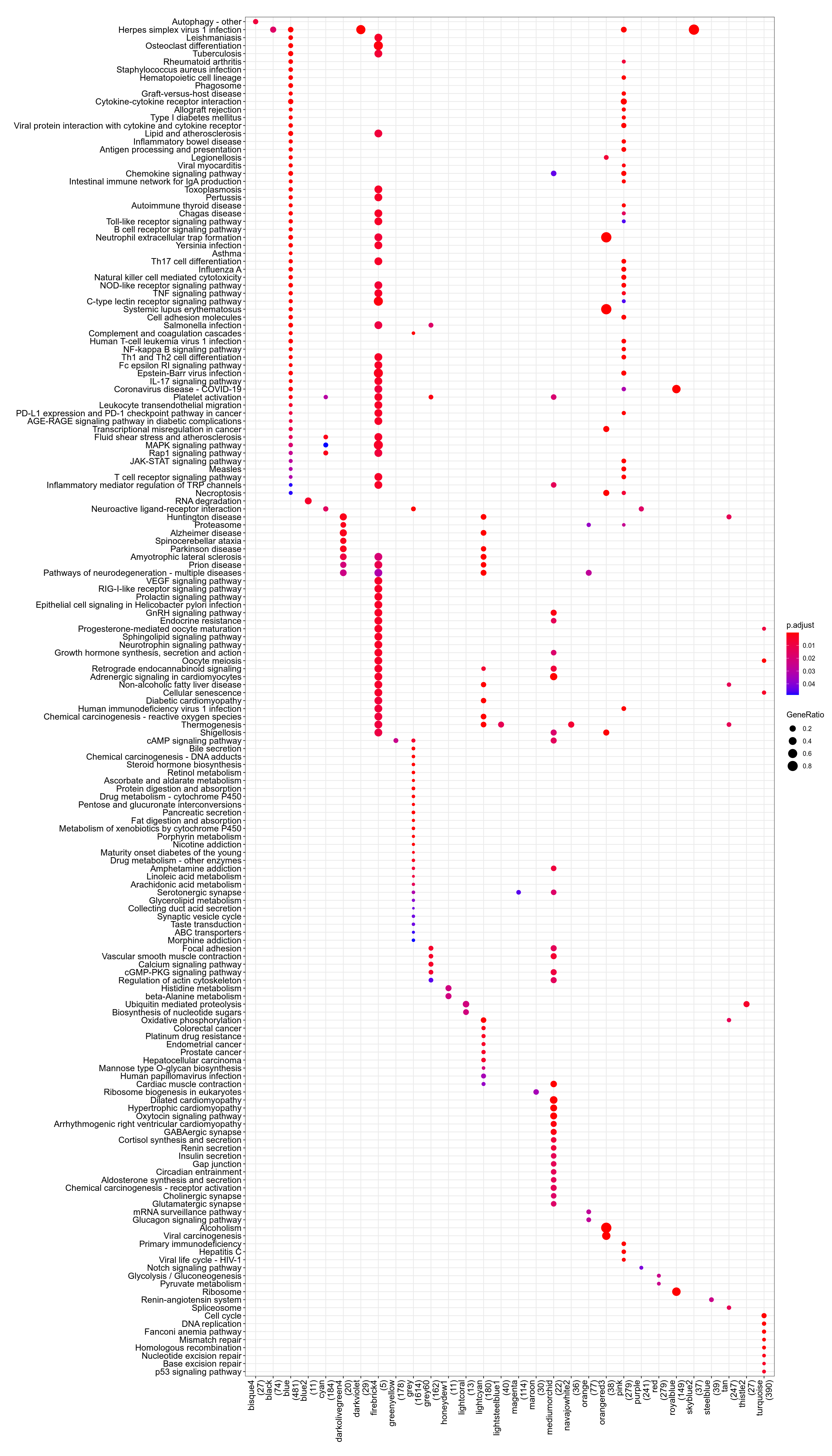
Figure 3.118: Kegg
.](_main_files/figure-html/resultsWgcnaTcgaSarcSTLMSKnitPlotEnrichHallmarks-1.png)
Figure 3.119: MSigDb Hallmarks. readable pdf version.
## No enriched DO term in these classesTo check that module-to-genes reattribution is working as expected, heatmap of signal for VNN1 module is produced, both as absolute values and as Z-score ones.
.](../../../out/bookdown/VNN1_sarcoma/plots/resultsWgcnaTcgaSarcSTLMSKnitPlotHeatmapVNN1ModuleAbsoluteValues.png)
Figure 3.120: log2(vst) signal in VNN1 module. Rows were gene name is prefixed with "-" display negative Z-score for these genes, meaning their signal is anticorrelated with other genes in the cluster. pdf version with readable genes and findable VNN1.
.](../../../out/bookdown/VNN1_sarcoma/plots/resultsWgcnaTcgaSarcSTLMSKnitPlotHeatmapVNN1ModuleZscoreValues.png)
Figure 3.121: Zscore of log2(vst) signal in VNN1 module. Rows were gene name is prefixed with "-" display negative Z-score for these genes, meaning their signal is anticorrelated with other genes in the cluster. pdf version with readable genes and findable VNN1.
.](_main_files/figure-html/resultsWgcnaTcgaSarcSTLMSKnitPlotGenesInVNN1ModuleCorPearson-1.png)
Figure 3.122: Genes in VNN1 module plotted by Pearson correlation against it. Details are available in this table.
- Enrichr results for all genes in VNN1 module are available here
- Enrichr results for genes in VNN1 module and positively correlated with it are available here
- Enrichr results for genes in VNN1 module and negatively correlated with it are available here
- g:Profiler results for all genes in VNN1 module are available here
- g:Profiler results for genes in VNN1 module and positively correlated with it are available here
- g:Profiler results for genes in VNN1 module and negatively correlated with it are available here
- g:Profiler results for comparison of the three queries above are available here