3.2.3.4 UPS
There are 36886 genes and 44 samples in the dataset.
At least some samples or genes look not suitable for the co-expression analysis. After removing them, there remain 36769 genes and 44 samples.
## Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
## 1 1 0.266 -2.47 0.782 5620.000 5.33e+03 9600.0
## 2 3 0.857 -2.68 0.967 430.000 3.29e+02 1930.0
## 3 5 0.872 -2.41 0.970 69.400 3.81e+01 659.0
## 4 7 0.876 -2.17 0.979 17.300 6.46e+00 285.0
## 5 9 0.900 -1.94 0.989 5.760 1.41e+00 141.0
## 6 11 0.913 -1.80 0.988 2.380 3.72e-01 84.1
## 7 13 0.914 -1.71 0.968 1.160 1.13e-01 59.2
## 8 15 0.937 -1.56 0.933 0.653 3.82e-02 43.6
## 9 17 0.877 -1.59 0.854 0.410 1.39e-02 40.7
## 10 19 0.389 -2.43 0.340 0.283 5.34e-03 39.9
## 11 21 0.402 -2.65 0.344 0.211 2.16e-03 39.4
## 12 23 0.416 -2.77 0.320 0.168 9.01e-04 39.0
## 13 25 0.424 -2.67 0.327 0.140 3.87e-04 38.8
## 14 27 0.430 -2.58 0.327 0.122 1.70e-04 38.6
## 15 29 0.436 -2.53 0.331 0.109 7.56e-05 38.5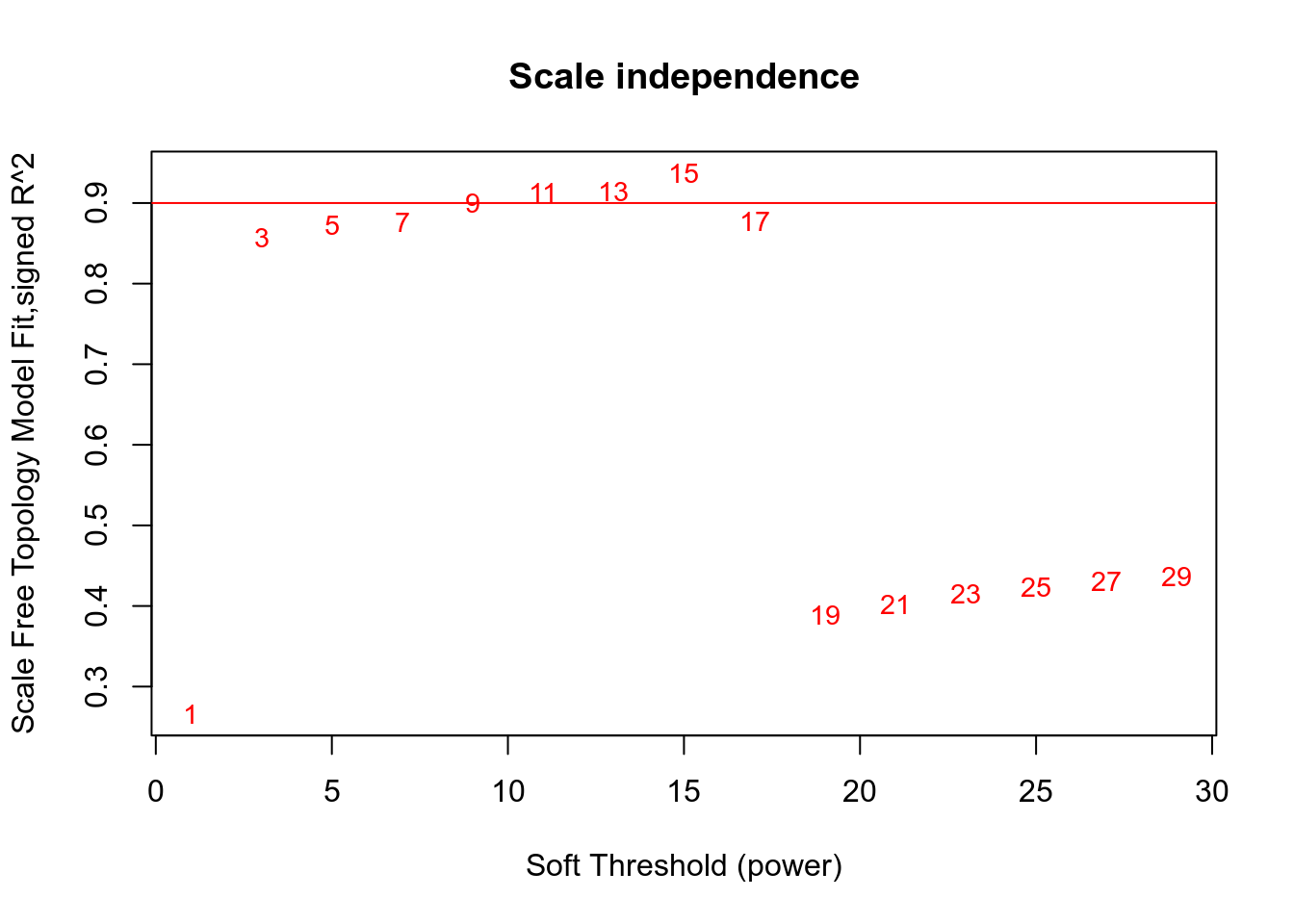
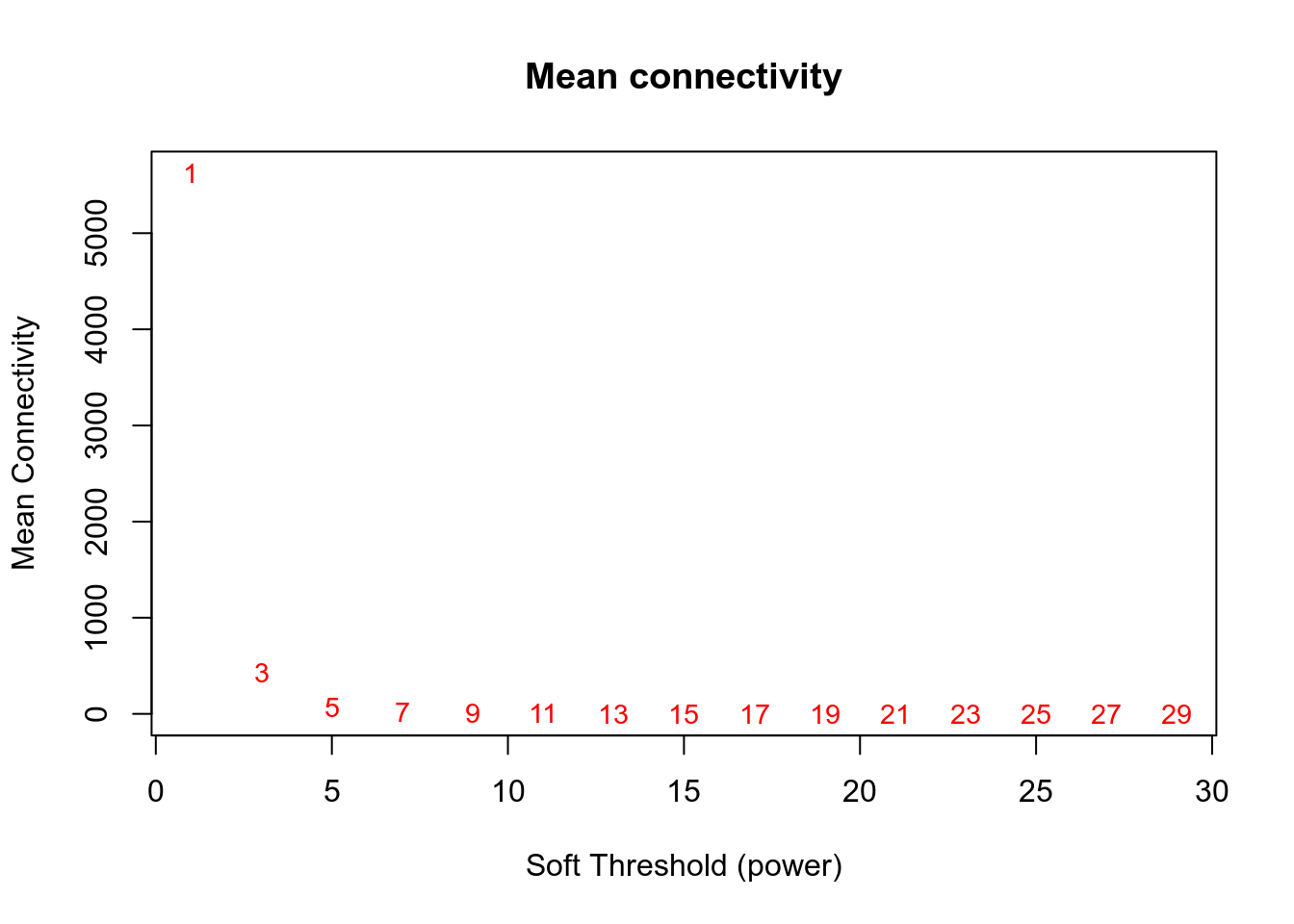
Network is computed using power=3.
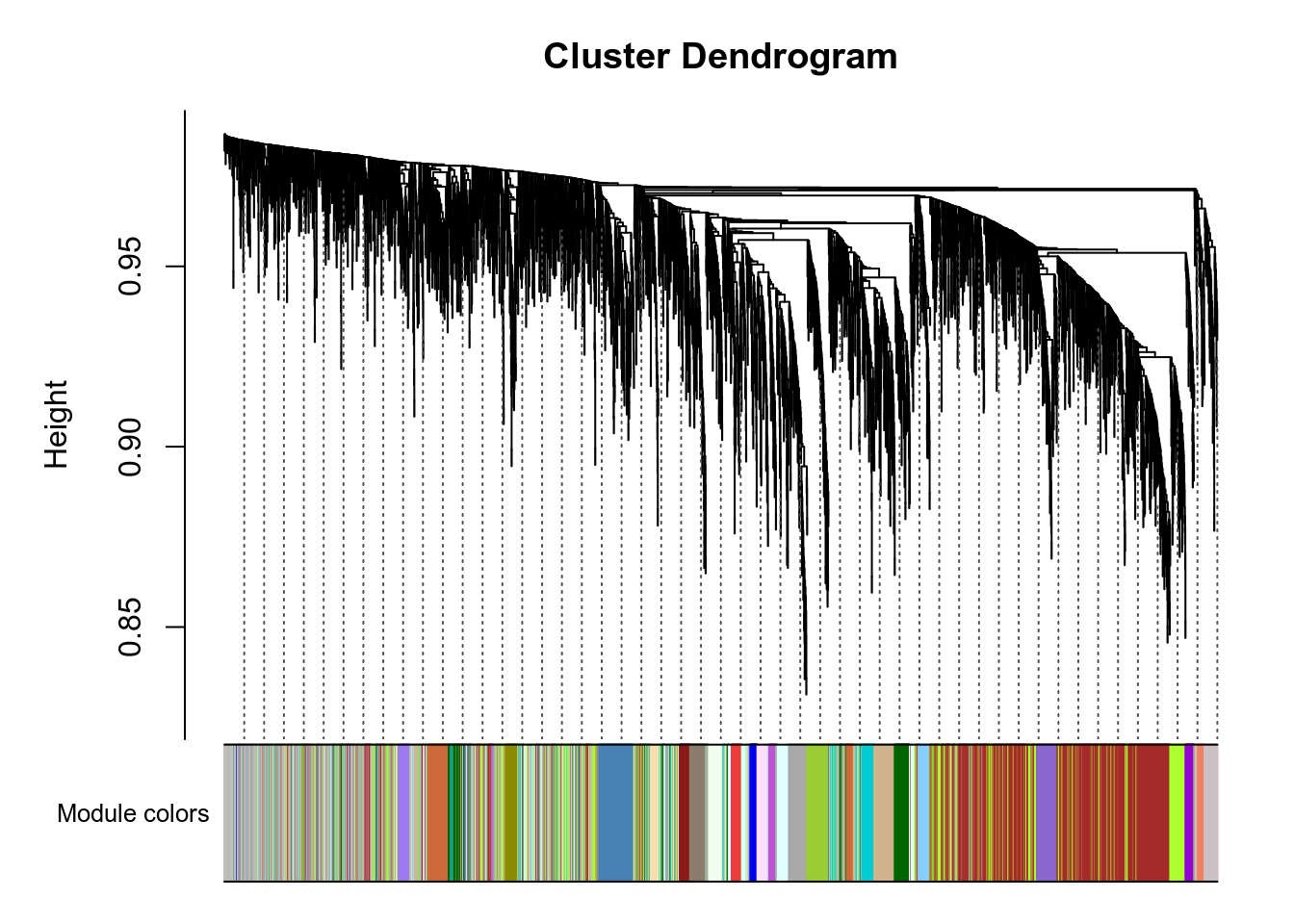
Here are the modules for a few genes of interest:
- VNN1 is in the blue module.
- PPARA is in the saddlebrown module.
- PPARD is in the grey module.
- PPARG is in the grey module.
- PPARGC1A is in the grey module.
- PPARGC1B is in the lightgreen module.
g:Profiler results for all modules are available here. Since there are many modules, you should expect at least 1 minute computation on the g:Profiler website for this query. Check below for faster queries focused on VNN1 module.
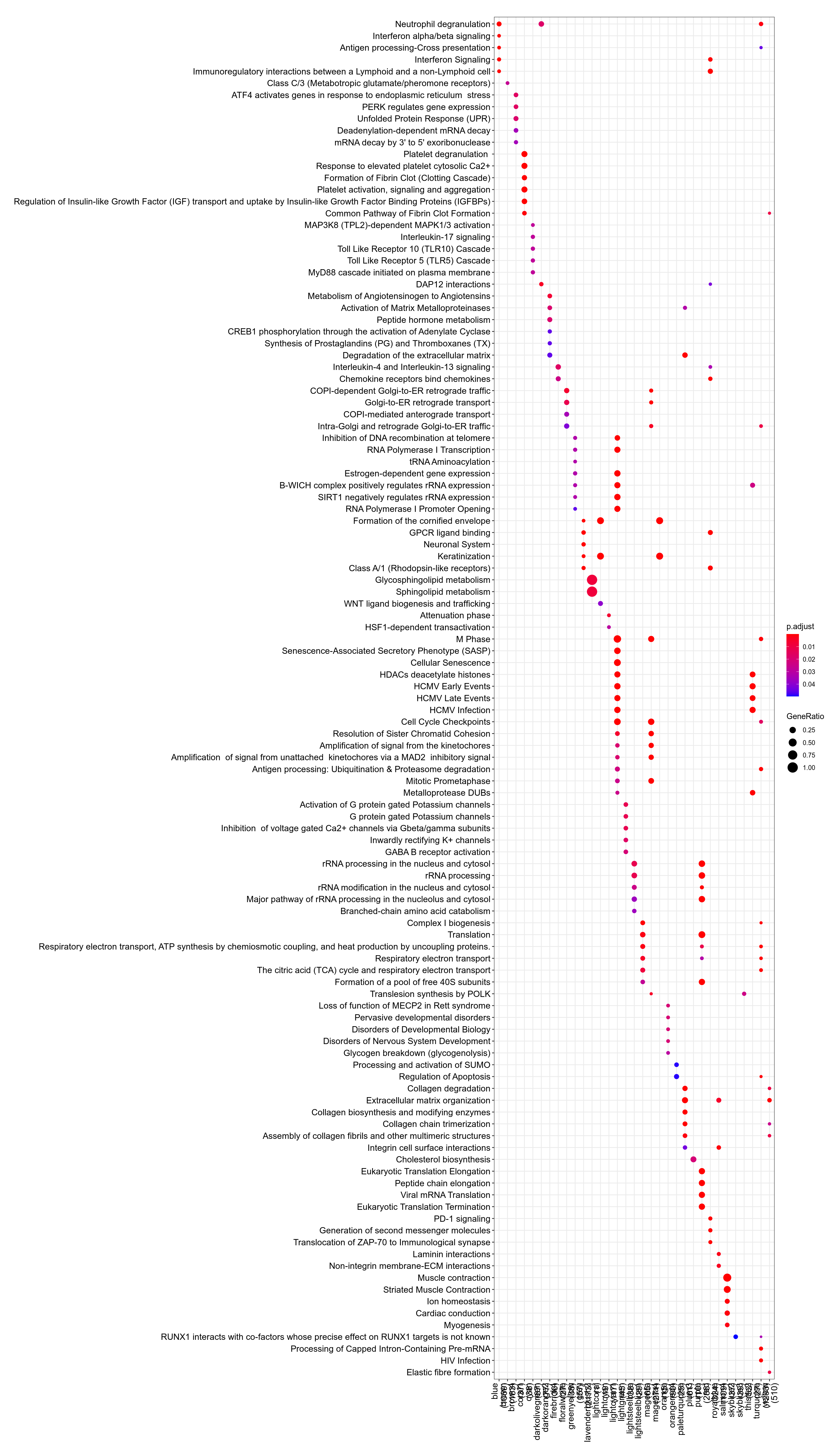
Figure 3.133: Pathway
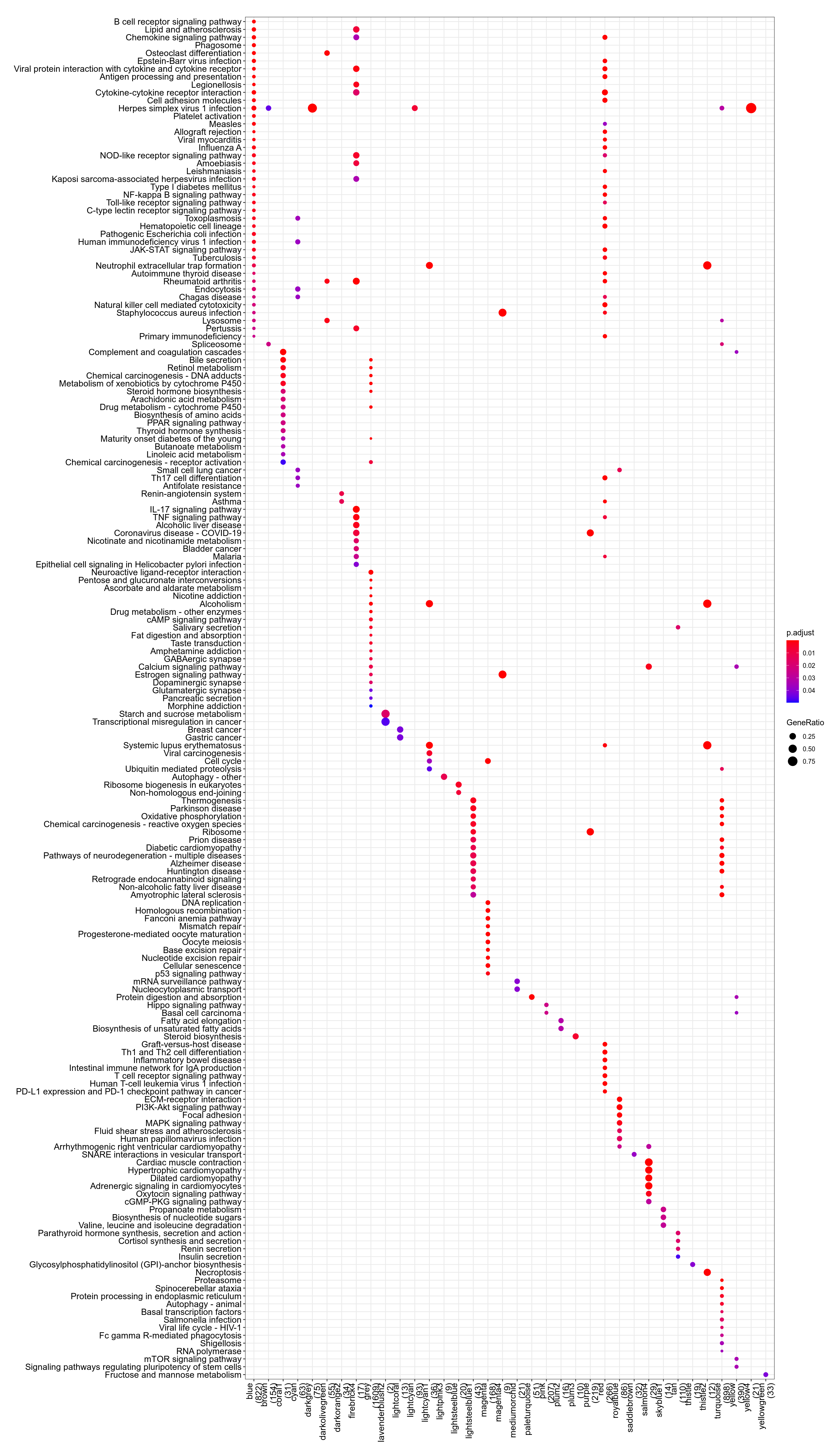
Figure 3.134: Kegg
.](_main_files/figure-html/resultsWgcnaTcgaSarcUPSKnitPlotEnrichHallmarks-1.png)
Figure 3.135: MSigDb Hallmarks. readable pdf version.
## No enriched DO term in these classesTo check that module-to-genes reattribution is working as expected, heatmap of signal for VNN1 module is produced, both as absolute values and as Z-score ones.
## VNN1 module contains more than 2000 genes. Heatmap generation is skippedVNN1 module contains more than 2000 genes. Heatmap generation is skipped
.](_main_files/figure-html/resultsWgcnaTcgaSarcUPSKnitPlotGenesInVNN1ModuleCorPearson-1.png)
Figure 3.136: Genes in VNN1 module plotted by Pearson correlation against it. Details are available in this table.
- Enrichr results for all genes in VNN1 module are available here
- Enrichr results for genes in VNN1 module and positively correlated with it are available here
- Enrichr results for genes in VNN1 module and negatively correlated with it are available here
- g:Profiler results for all genes in VNN1 module are available here
- g:Profiler results for genes in VNN1 module and positively correlated with it are available here
- g:Profiler results for genes in VNN1 module and negatively correlated with it are available here
- g:Profiler results for comparison of the three queries above are available here